Projects
Explore my data science and development projects
MET Art Display Predictor
ML-powered prediction of artwork exhibition likelihood - using data from the Metropolitan Museum of Art
Analysis of public data from the Metropolitan Museum of Art and a gradient boosting algorithm predicting which art piece is on view
This project uses data made available by The Metropolitan Museum of Art under the Open Access program.
MET Open Access Dataset: https://github.com/metmuseum/openaccess
Try the Model
Dataset Analysis
Class Distribution
Model Performance Dashboard
Master's Thesis - Modality fusion in enzyme interaction prediction
Modality-fusion strategies for a transformer predicting enzyme-substrate interactions
- Constructed a novel dataset of ~185,000 data points by mapping protein sequences to 3D structures from AlphaFold using the Graphein library.
- Represented protein structures as graphs and generated node embeddings using the Node2Vec algorithm.
- Designed & implemented novel modality fusion techniques (additive & multiplicative) to merge protein sequence and structural embeddings before feeding them into a pre-trained transformer, avoiding costly retraining from scratch.
- Trained and evaluated model performance end-to-end, including a gradient boosting (XGBoost) ensemble step, across various data split scenarios (random, cold protein, cold SMILES).
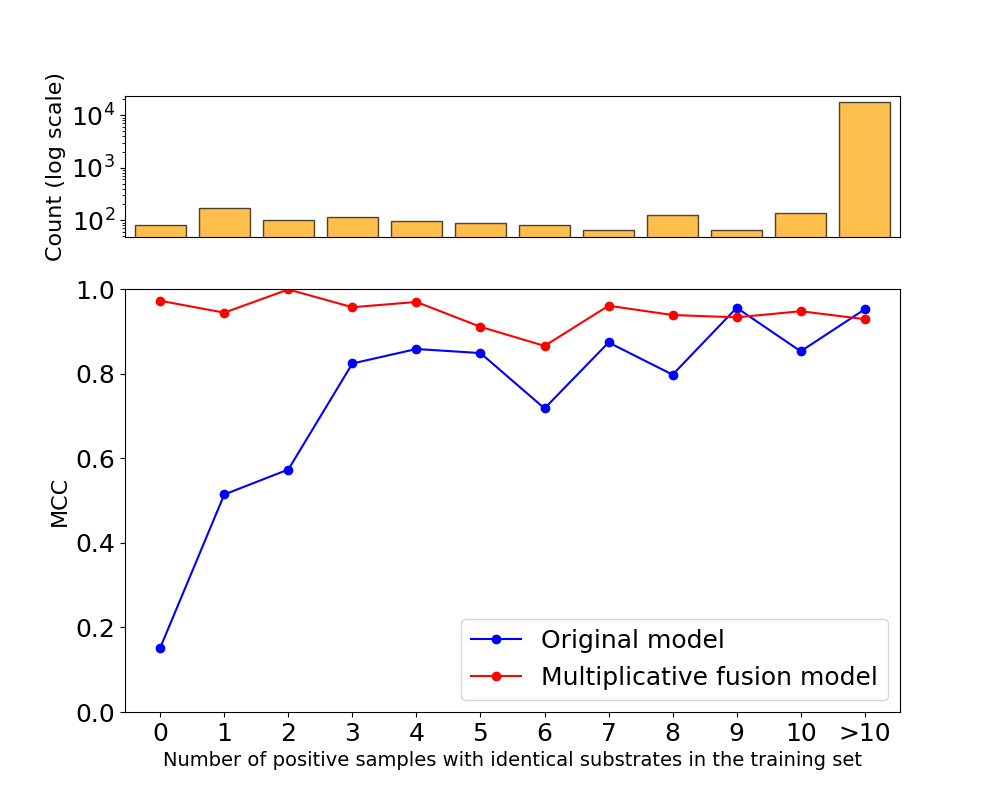
- Validated that the model maintains high performance (~97% accuracy) on standard random splits.
- Demonstrated improved generalization, achieving a ~1% accuracy increase on challenging "cold start" splits with unseen small molecules—the primary failure mode of the original model.
- Showcased significantly better performance (via MCC score) for rare, underrepresented substrates, highlighting the model's potential for novel discovery.
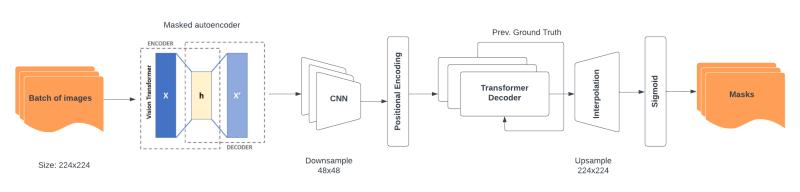
Tranformer-based Lane Detection
Transformer-based lane detection that leverages spatiotemporal information from sequential video frames to robustly dynamic road conditions
- Curated and preprocessed large-scale driving datasets (CULane: 55h / 133k frames, TuSimple), converting lane point annotations into segmentation masks.
- Designed and applied a data augmentation pipeline (random flips, rotations ±30°, crops, and normalization) to improve robustness to camera pose changes, occlusions, and viewpoint shifts.
- Built a transformer-based segmentation pipeline that leverages temporal information across video frames to improve lane detection consistency.
- Evaluated and replaced a ResNet34 backbone with a self-supervised MAE-ViT, validating superior lane-specific feature extraction via custom image-reconstruction experiments.
- Demonstrated MAE-ViT's advantage over CNN backbones for capturing long, thin, and partially occluded lane structures.
- Showed that temporal modeling with transformers stabilizes training and improves qualitative lane predictions under challenging conditions.
- Delivered a validated proof-of-concept under strict hardware constraints.
Lindel
Reproducing, Evaluating and Interpreting the Lindel Model for CRISPR-Cas9 Repair Outcome Prediction
🧬 Project Overview
In a collaborative academic project, our team reproduced and validated key findings from Chen et al.’s seminal work on predicting CRISPR-Cas9 DNA repair outcomes. We successfully replicated the core functionality of their machine learning model, Lindel, confirming both its superiority over a baseline model and the high reproducibility of mutational outcomes across experiments.
🔍 Individual Research Focus
My individual contribution centered on interpreting the model’s internal logic to critically evaluate the authors’ claims regarding sequence context importance.
🎯 Objective
Assess whether the logistic regression model used to predict the insertion–deletion (indel) ratio learns the same sequence context importance as reported in the original paper.
🛠️ Methods
To interpret and validate the model’s behavior, I applied a complementary set of interpretability techniques:
- Learned weights analysis of the neural network
- SHAP (SHapley Additive exPlanations) for feature importance
- Multiple Correspondence Analysis (MCA) to examine relationships in categorical sequence data
✅ Key Findings
- Confirmed the strong influence of a single nucleotide (‘T’) at the cut site (position 17),
though its impact on model predictions differed from the paper’s biochemical interpretation. - Questioned the reported importance of specific dinucleotides (e.g., TG, CG, GA),
as their effects were not consistently supported across interpretability analyses. - Concluded that the model relies more heavily on single-nucleotide features than originally proposed,
offering a more nuanced and critical understanding of the model’s decision-making process.